New!! Full-length 16S rRNA sequencing
The Microbiome Core now offers full-length 16S sequencing on the PacBio Revio for higher accuracy and better taxonomic resolution, supporting species level identification for your microbial community studies. Contact us for more information.
We provide microbiome services, beginning with study design through statistical analysis of data obtained from various microbial assays. We have well-established protocols for DNA extractions from diverse sample types and amplicon analysis for 16S and 18S (Illumina MiSeq), as well as full-length analysis of 16S and ITS (PacBio Revio). All procedures are performed in dedicated hoods and with optimized protocols to minimize the possibility of background or cross contamination during sample processing.

Project management
- SOPs for all processes
- study design
- sample tracking
Aliquoting samples (fecal, oral)
- aliquot frozen aliquots into cryovials
- maintain, organize and QC inventory of samples for project
Nucleic Acid extraction
- Qiagen DNA kits (Powersoil Pro)
- Qiacube HT automated extraction
- ZymoBIOMICS DNA Miniprep
- others available upon request
Sample Quality Assessment and Shearing
- Qubit
- Agilent Bioanalyzer 2100 assays
- Nanodrop
Microbiome Profiling
- 16S sequencing via Earth Microbiome Project protocols for V4, V1-V3
- 18S sequencing via Earth Microbiome Project protocols
- ITS sequencing via Earth Microbiome Project protocols
- PacBio Kinnex full-length16S sequencing
- PacBio Kinnex full-length ITS sequencing
- Metagenomic sequencing, visit Sequencing and Genomic Technologies Core
Agilent Bioanalyzer

The Agilent Bioanalyzer DNA assay leverages microfluidics technology, enabling the quality analysis of DNA using only 1ul of sample. The assay is run on the Agilent 2100 Bioanalyzer instrument and utilizes the Agilent 2100 Expert Software to analyze and display results. RNA and DNA fragment sizes and concentrations are generated for each sample.
Please use the guidelines below to dilute your samples to the appropriate range BEFORE submitting them for Quality Check. Also, please provide 4ul of sample for quality check. DNA must be stored at -20C and RNA must be stored at -80C.
Assay types and information:
- DNA Analysis Kits and Reagents: Details and Specifications
- Agilent High Sensitiviy DNA
- Checking your Agilent High Sensitivity DNA Assay Results
- RNA Analysis Kits
Qubit Fluorometer
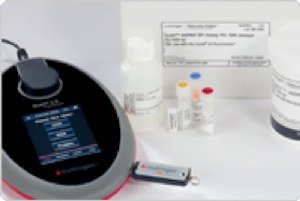
The Qubit 2.0 Fluorometer quantitates DNA, RNA, and protein with unprecedented accuracy, sensitivity, and simplicity. It is designed for molecular biology labs that work with precious samples that are rare or difficult to process and applications requiring precise measurement, such as real-time PCR.
| Assay Type | Size Range | Concentration Range | Max # of samples per chip |
| Bioanalyzer High Sensitivity DNA | 50 - 7000 bp | 0.005 - 0.5 ng/ul | 11 |
| Qubit dsDNA HS | any | 0.001 – 0.5ng/ul | NA |
| Qubit dsDNA BR | any | 0.1 - 1000 ng/ul | NA |

Promega Glomax Discover
Promega GloMax® Discover is a ready-to-use multimode plate reader developed with Promega reagent chemistries to provide a simple means of detecting luminescence, fluorescence and absorbance. This advanced plate reader provides flexible use of filters and includes high-performance luminescence, fluorescence, UV-Visible absorbance, BRET and FRET, two-color filtered luminescence and kinetic measurement capabilities.
Data Analysis and Computational Resources:
Packages for standard analysis of 16S and 18S amplicon or full-length sequencing data are available through the core. Additional custom and statistical analysis, including correlation and differential abundance, are available on an hourly consultation basis.
For specific analysis questions and the opportunity to interact with your Microbiome colleagues, the Duke Microbiome Center also hosts biweekly Microbiome office hours and a bioinformatics/biostatistics working group.
Data Management:
Data will be returned to the investigator by Globus (NGS results from Duke Sequencing and Genomic Technologies Core) or through DukeBox (all other data). Instructions will be sent to investigator on how to download the data once results are posted. Please note that all investigators will be responsible for long term storage and sharing of data generated for their projects. If you would like assistance with depositing data into the SRA database, please contact us.
Please contact us for the most up-to-date rates at microbiomesr@duke.edu.