
Associate Professor in Integrative Immunobiology
Associate Professor of Cell Biology
Member of the Duke Human Vaccine Institute
Member of the Duke Cancer Institute
Box 3054
Durham, NC 27710
Phone: (919) 684-1351
Fax: (919) 684-2790
nicholas.heaton@dm.duke.edu
Research
Our research group is primarily interested in the study of respiratory RNA viruses. We have published work on viruses of the families: orthomyxoviridae, paramyxoviridae, and coronaviridae. In particular, our current work is focused on the orthomyxovirus influenza A, as well as the coronavirus SARS-CoV-2, as these are the viruses that have caused, and remain capable of causing, global pandemics.
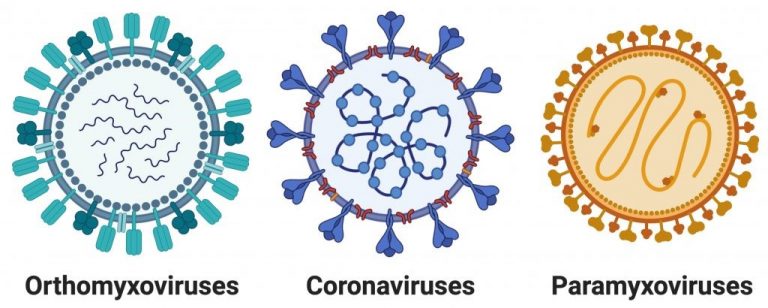
Influenza viruses are negative sense, RNA viruses that encode their genome across eight genomic segments. Due to the segmented nature of their genome, when a cell is infected with multiple viruses, segment exchange can lead to the generation of new viral variants. These “reassorted” viruses, frequently after reassortment between human and avian/swine strains of influenza, have the ability to cause global pandemics.
Coronaviruses are a family of positive sense, RNA viruses which are known to infect a large range of animals. While there are a number of human coronaviruses, epidemic or pandemic outbreaks are primarily thought to occur when a virus “jumps” from an animal to a human host. The research in our laboratory is currently focused on three major areas of investigation, all with the ultimate goal of developing new therapies that can be used to combat both current and future influenza and coronavirus outbreaks.
Our main research areas are:
1.) Understanding how viral infection induces lung inflammation, and how that inflammation influences recovery from disease.
One of the experimental approaches that the lab frequently takes is to develop new viral-based research tools. These reporter viruses, together with transgenic animals, have allowed us to ask questions about the biology of viral infections that would have been difficult or impossible to ask with unmodified viruses. In one of our favorite approaches, we have utilized recombinase expressing viruses to permanently label virally infected cells.
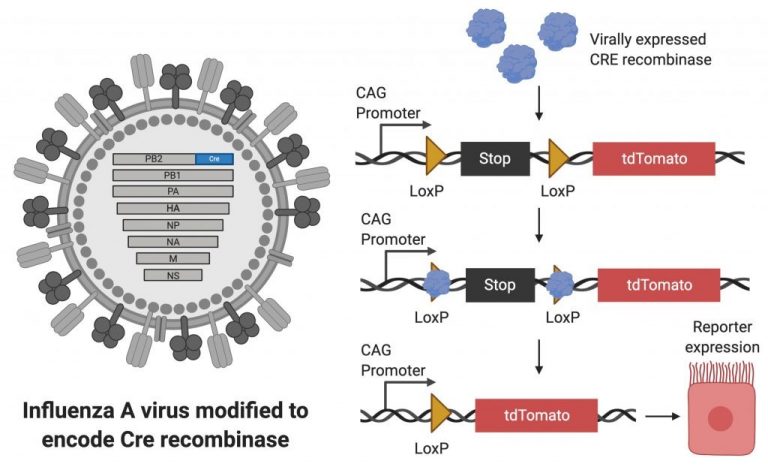
This approach has allowed us to understand how pulmonary epithelial cells are changed by viral infection. We have found that many of these changes influence inflammation in the lung, which affects both the severity of viral disease and the time to recovery. Continued work in this area may eventually lead to therapies that help improve recovery rates after viral pneumonia.
2.) Developing biologics and vaccines to prevent viral disease.
We have also developed virion-based platforms that can be used to generate next-generation anti-viral biologics or vaccines that avoid some of the problems associated with current influenza intervention strategies and vaccines.
>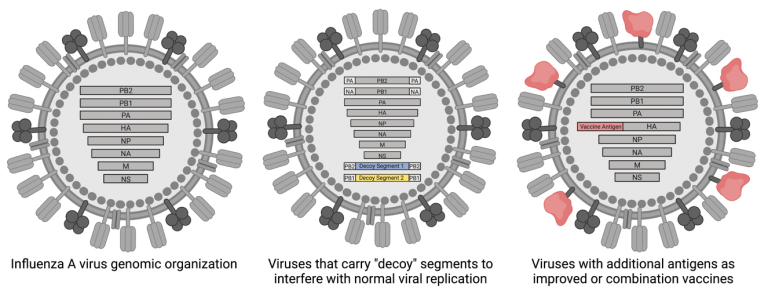
For example, by generating viral particles that carry “decoy” genomic segments, we can help prevent viral reassortment and spread. An additional way that we have been working to improve influenza vaccines is the incorporation of additional viral antigens into standard vaccine backgrounds/platforms. Approaches like this may allow us to generate a broader, and ultimately more protective, response to vaccination.
3.) Defining the host proteins that are either required for, or can restrict, viral infection of airway epithelial cells.
Human viruses have dramatically less genetic material relative to their hosts; they therefore rely on co-opting host proteins to complete their replication cycles. Additionally, humans have evolved defensive proteins, that when expressed, are capable of restricting viral infection and/or spread. It isn’t fully understood however, which proteins fall into these categories. Our lab has utilized high throughput screening approaches to define host proteins that fall into both categories.
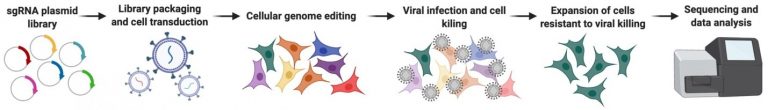
Our goal is to not only identify these key host proteins, but also develop ways to manipulate them (either decrease expression of required factors or over-express restriction factors) in order to develop new antiviral therapeutics. Further, since many viruses utilize the same host factors, these types of studies may lead to the identification of broadly-acting therapeutics.
Altogether, we take a number of complementary research approaches in order to develop new ways to combat respiratory viral disease. There are many individual projects within these research areas, and we are always looking for new people to join our team. Please get in touch if you are interested in working with us or learning more!
Biography
Dr. Heaton received BS degrees in Bacteriology and Biology from the University of Wisconsin-Madison in 2007. He then did his graduate studies in the laboratory of Dr. Glenn Randall at the University of Chicago working on the roles of lipid metabolism during positive stranded RNA virus replication. He received his PhD in 2012.
For postdoctoral studies, Dr. Heaton joined the laboratory of Dr. Peter Palese to study the molecular biology of influenza viruses in 2012. Dr. Heaton joined the faculty of the Molecular Genetics and Microbiology department at Duke University Medical Center in 2015. His laboratory is currently focused on developing reporter viruses for use in conjunction with transgenic animals to understand the impact of infected cell fates on lung disease and immunity.
Lab Members
Publications
Recent, selected publications from the lab:
- Trimarco JD, Nelson SL, Chaparian RR, Wells AI, Murray NB, Azadi P, Coyne CB, Heaton NS. Cellular glycan modification by B3GAT1 broadly restricts influenza virus infection. Nat Commun. 2022 Oct 29;13(1):6456. doi: 10.1038/s41467-022-34111-0. PMID: 36309510; PMCID: PMC9617049.
- Yaron TM, Heaton BE, Levy TM, Johnson JL, Jordan TX, Cohen BM, Kerelsky A, Lin TY, Liberatore KM, Bulaon DK, Van Nest SJ, Koundouros N, Kastenhuber ER, Mercadante MN, Shobana-Ganesh K, He L, Schwartz RE, Chen S, Weinstein H, Elemento O, Piskounova E, Nilsson-Payant BE, Lee G, Trimarco JD, Burke KN, Hamele CE, Chaparian RR, Harding AT, Tata A, Zhu X, Tata PR, Smith CM, Possemato AP, Tkachev SL, Hornbeck PV, Beausoleil SA, Anand SK, Aguet F, Getz G, Davidson AD, Heesom K, Kavanagh-Williamson M, Matthews DA, tenOever BR, Cantley LC, Blenis J, Heaton NS. Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication. Sci Signal. 2022 Oct 25;15(757):eabm0808. doi: 10.1126/scisignal.abm0808. Epub 2022 Oct 25. PMID: 36282911.
- Zhu X, Trimarco JD, Williams CA, Barrera A, Reddy TE, Heaton NS. ZBTB7A promotes virus-host homeostasis during human coronavirus 229E infection. Cell Rep. 2022 Oct 25;41(4):111540. doi: 10.1016/j.celrep.2022.111540. Epub 2022 Oct 5. PMID: 36243002; PMCID: PMC9533670.
- Chaparian RR, Harding AT, Hamele CE, Riebe K, Karlsson A, Sempowski GD, Heaton NS*, Heaton BE*. A Virion-Based Combination Vaccine Protects against Influenza and SARS-CoV-2 Disease in Mice. J Virol. 2022 Jul 12:e0068922. doi: 10.1128/jvi.00689-22. Epub ahead of print. PMID: 35862698. *Co-corresponding authors
- Froggatt HM, Burke KN, Chaparian RR, Miranda HA, Zhu X, Chambers BS, Heaton NS. Influenza A virus segments five and six can harbor artificial introns allowing expanded coding capacity. PLoS Pathog. 2021 Sep 27;17(9):e1009951. doi: 10.1371/journal.ppat.1009951. Epub ahead of print. PMID: 34570829.
- Froggatt HM, Harding AT, Chaparian RR, Heaton NS. ETV7 limits antiviral gene expression and control of influenza viruses. Sci Signal. 2021 Jul 13;14(691):eabe1194. doi: 10.1126/scisignal.abe1194. PMID: 34257104
- Trimarco JD, Heaton BE, Chaparian RR, Burke KN, Binder RA, Gray GC, Smith CM, Menachery VD, Heaton NS. TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog. 2021 May 27;17(5):e1009599. doi: 10.1371/journal.ppat.1009599. PMID: 34043740; PMCID: PMC8189496.
- Luo Z, Girton AW, Heaton BE, Heaton NS. Engineered influenza virions reveal the contributions of non-hemagglutinin structural proteins to vaccine mediated protection. J Virol. 2021 Mar 3;95(10):e02021-20. doi: 10.1128/JVI.02021-20. Epub ahead of print. PMID: 33658342; PMCID: PMC8139674.
- Harding AT, Goff MA, Froggatt HM, Lim JK, Heaton NS. GPER1 is required to protect fetal health from maternal inflammation. Science. 2021 Jan 15;371(6526):271-276. doi: 10.1126/science.aba9001. PMID: 33446553; PMCID: PMC8060949.
- Froggatt HM, Heaton BE, Heaton NS. Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLproReporter Assay. Journal of Virology. 2020 Oct 27;94(22):e01265-20. doi: 10.1128/JVI.01265-20. PMID: 32843534; PMCID: PMC7592234.
- Dumm RE, Wellford SA, Moseman EA*, Heaton NS*. Heterogeneity of Antiviral Responses in the Upper Respiratory Tract Mediates Differential Non-lytic Clearance of Influenza Viruses. Cell Reports. 2020 Sep 1;32(9):108103. doi: 10.1016/j.celrep.2020.108103. PMID: 32877682; PMCID: PMC7462569. *Co-corresponding authors
- Harding AT, Haas GD, Chambers BC, Heaton NS. Influenza viruses that required 10 genomic segments as antiviral therapeutics. PLoS Pathogens. 2019. Nov 15;15(11):e1008098. doi: 10.1371/journal.ppat.1008098. PMID: 31730644
- Chambers BS, Heaton BE, Rausch K, Dumm RE, Hamilton JR, Cherry S*, Heaton NS*. DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection. Nature Microbiology. 2019. Nov;4(11):1964-1977. doi: 10.1038/s41564-019-0509-3. PMID: 31358986 *Co-corresponding authors
- Dumm RE, Fiege JK, Waring BM, Kuo CT, Langlois RA, Heaton NS. Non-lytic clearance of influenza B virus from infected cells preserves epithelial barrier function. Nature Communications. 2019 Feb 15;10(1):779. doi: 10.1038/s41467-019-08617-z. PMID: 30770807
- Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, Heaton NS. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Reports. 2017 20(7): p1503-1512.
- Harding AT, Heaton BE, Dumm RE, Heaton NS. Rationally Designed Influenza Virus Vaccines That Are Antigenically Stable during Growth in Eggs. mBio. 2017 June 6, Vol. 8 no.3 e00669-17 doi: 10.1128/mBio.00669-17. PMID: 28588131
- Hamilton JR, Sachs D, Lim JK, Langlois RA, Palese P, Heaton NS. Club cells surviving influenza A virus infection induce temporary non-specific anti-viral immunity. Proc Natl. Acad Sci USA. 2016 April 5, vol. 113 no. 14, 3861-3866. doi: 10.1073/pnas.1522376113. PMID: 27001854
- Fulton BO, Palese P, Heaton NS. Replication competent influenza B reporter viruses as tools for screening antivirals and antibodies. Journal of Virology. 2015 Dec 1;89(23):12226-31. doi: 10.1128/JVI.02164-15. Epub 2015 Sep 23. PMID:26401044
- Fulton BO, Sachs D, Beaty SM, Won ST, Lee B, Palese P, Heaton NS. Mutational analysis of measles virus suggests constraints on antigenic variation of the glycoproteins. Cell Reports. 2015 Jun 9;11(9):1331-8. doi: 10.1016/j.celrep.2015.04.054. Epub 2015 May 21. PMID:26004185
For a complete list of publications from Dr. Heaton, please click here.





