Research
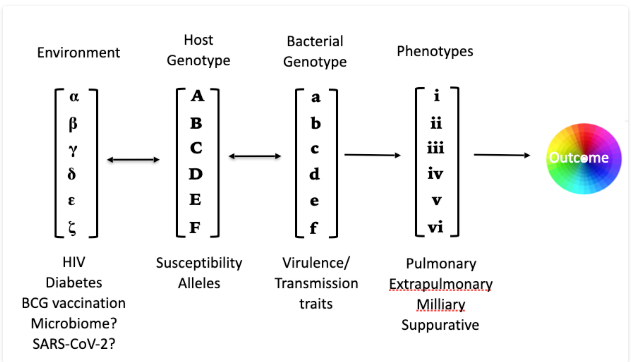
The Smith Lab are interested in host genetic diversity, bacterial variation, and how these host-pathogen genetic interactions drive tuberculosis disease states.
Systems Genetics of Tuberculosis: We leverage host diversity in mice and macrophages from wild-derived mouse strains and diverse mouse panels, including the Collaborative Cross and BXD mammalian resources. In parallel, we define the bacterial genetic requirements for growth and adaptation across these diverse host environments through new mycobacterial genetic approaches, including saturated libraries of transposon mutants (TnSeq), CRISPRi and knockouts generated through ORBIT (“oligonucleotide-mediated recombineering followed by Bxb1 integrase targeting”). These combined host and bacterial genome-wide approaches allows the interrogation of each host-pathogen interaction underlying TB.
Want to learn more?
Check out how we combine diverse Mammalian and Mycobacterial genetics to map the host-interacting with pathogen loci (hipQTL) across the genome. Read about our new hipQTL preprint here!
Leveraging the Collaborative Cross mouse panel we have identified new models of distinct TB disease states and protective immunity. Check out how we identified a causal host variant here.
We leverage this dual host-pathogen system to understand how vaccines alter disease outcome in diverse hosts. Read about how host genetic background underlies BCG vaccine efficacy here.
Biography
Clare has a Bachelor of Biotechnology with first-class honors from the University of Tasmania in Australia, where she discovered the world of host genetics and disease susceptibility. She then got her PhD at the Menzies Research Institute under the mentorship of geneticist Prof Simon Foote. Here she identified a red cell factor essential for growth of the malarial parasite, Plasmodium falciparum. She found that red cells from people or mice with deficiencies in haem biosynthetic enzymes were more resistant to malarial growth, and this protective effect could be mimicked with drugs as a new host-directed antimalarial therapy.
Clare did her postdoc in the lab of Prof Christopher Sassetti at Umass Medical School in MA. Here, she combined her host genetics background with new bacterial technologies to develop a “dual genome” system to probe the host-pathogen interactions that drive susceptibility to Tuberculosis. By combining transposon libraries of Mycobacterium tuberculosis with genetically diverse mice, including the Collaborative Cross, she defined the immunological niche and conditions that Mtb requires to grow in genetically diverse hosts. She additionally established the Collaborative Cross as a new model to dissect vaccine protection and TB pathogenesis across diverse TB clinical isolates.
Now, as an Assistant Professor and newest recruit to the Molecular Genetics and Microbiology (MGM) department at Duke, she is bringing systems genetics approaches to define the host-pathogen interactions that drive susceptibility to tuberculosis. By understanding how these interactions drive immunity, new host-pathogen paired therapeutics and vaccines can be rationally designed. She is excited to help train the next-gen of researchers that will help solve the big questions in medical research.


