Research
Human genetic variation regulating infection and inflammation
Despite improvements in public health, advancements in vaccines, and the development of many classes of antibiotics, infectious disease is still responsible for over a quarter of all deaths worldwide. However, even for the most devastating of pandemics, individuals demonstrate a large variability in the severity of infection. The long-term goal of the lab is to understand the genetic basis for differences in susceptibility to infection and related inflammatory disorders. We approach this question through a combination of experimental and computational approaches that combine high-throughput cell biology with quantitative human genetics. The identified genetic differences serve as the starting point for exploring new cell biology and human disease susceptibility genes.
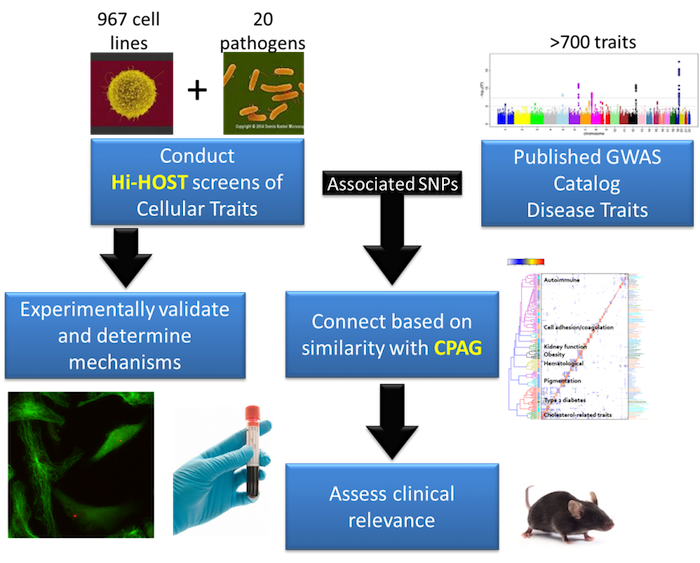
Using hundreds of genotyped cell lines from different people, we measure inter-individual variation in host-pathogen traits and identify associated genetic differences with a cellular genome-wide association study (GWAS) approach called Hi-HOST (High-throughput Human in vitrO Susceptibility Testing). Importantly, Hi-HOST serves as a platform for both identification of SNPs associated with cellular traits and experimental dissection of how they affect those traits. To connect these cellular traits to human disease susceptibility, we have developed a computational framework called CPAG (Cross-Phenotype Analysis of GWAS) to identify cellular traits that can explain a portion of the genetic risk for human diseases based on similarity of GWAS hits. In this way, genetic variants that modulate complex human diseases can be connected to experimentally tractable cellular models to study mechanisms. Using this experimental and analytic platform, the ultimate goal is to reveal critical genes and pathways involved in the pathophysiology of disease to enable development of new therapeutic targets.
We have accumulated screening data for nine different pathogens, which are now being studied to reveal basic mechanisms of host-pathogen interactions. Current research efforts are focused on genetic variants that impact:
- Invasion of cells by Salmonellato reveal basic mechanisms of cellular uptake and relevance to risk of typhoid fever
- Inflammatory cell death (pyroptosis) in response to Salmonellaand development of metabolites as biomarkers for sepsis outcomes
- Cytokine response to Salmonellato elucidate novel host-pathogen signaling pathways and their relevance to risk and severity of infection and autoimmunity
- Chlamydia trachomatis invasion, replication, and cytokine-induction as a means of understanding susceptibility and severity of this common sexually transmitted infection
- Intracellular replication of Yersinia pestis, one of the most feared pathogens in human history, which we suspect has left lasting evolutionary consequences in humans
In characterizing variation in these traits, the first goal is to understand the molecular/cellular mechanism of how the identified genetic variants intersect with known pathways. The second goal is to use our findings to guide clinical association studies and experiments with animal models to determine the relevance in disease. Thus our lab employs experimental, computational, and translational approaches to understand the consequences of human genetic variation and welcomes enthusiastic individuals interested in any or all of these approaches. We hope that the knowledge gained in these studies will help explain why some individuals are resistant to different infections and in developing therapies to decrease the mortality and morbidity of susceptible individuals.
Biography
Dennis Ko received a B.S. from Cornell University in 1997. His research career began in the lab of Dr. Jeffrey W. Roberts at Cornell University. Using a genetic screen of the bacterial initiation factor σ70 followed by biochemical characterization, he identified a surface on the protein important for transcriptional regulation by the phage Q protein and revealed a novel role for a sigma factor beyond transcription initiation.
His training continued from 1997-2005 in the MSTP at Stanford University. He completed both MD and PhD degrees but clearly saw that his future was in research aimed at understanding the biology of disease. His thesis project was carried out in the laboratory of Dr. Matthew P. Scott. Using a combination of biochemistry, cell biology, genetics and animal models, he pioneered a new subject in the lab examining how genetic alterations lead to the neurodegenerative lipid disorder, Niemann-Pick type C (NPC). The main findings of his studies on NPC were that: 1) mutation of NPC1 slowed the dynamic trafficking of the organelles in which it is found, contributing to the lipid derangements seen in this disorder; 2) NPC2 binds cholesterol with sub-micromolar affinity, and this binding is essential for activity; and 3) the requirement of NPC1 in the brain is cell autonomous, and its loss can lead to neuronal cell death with properties consistent with autophagy.
He took a year off for personal reasons following the completion of his MD/PhD degrees in June 2005. During this time, he volunteered in two labs on an informal basis to remain scientifically engaged. It was during this time that he decided to switch fields. Two major developments helped shape this decision: 1) the massive amount of genetic information from studies of human diversity and 2) improvements in studying mammalian cell biology, especially RNA interference and fluorescence assays. He, therefore, decided to engage in studies that would take advantage of these new developments in the field of host-pathogen interactions.
As a post-doctoral Life Sciences Research Foundation fellow in the lab of Samuel Miller at the University of Washington, Dr. Ko developed a novel screening method termed Hi-HOST (high throughput human in vitro susceptibility testing) for identifying human genetic variation that affects cell-based readouts of bacterial infection. Using this approach, he measured cellular variation in the human cell death response to Salmonella and identified common genetic differences that influence this in vitro trait. Through this approach, he was able to both discover unexpected cell biology and identify genetic differences important for human health and disease.
Dr. Ko joined the faculty of Duke Molecular Genetics & Microbiology, Medicine, and the Center for Human Genome Variation in 2012. His laboratory will continue applying the Hi-HOST approach to host-pathogen traits for Salmonellae, Yersinia, and other cellular phenotypes of infection and inflammation. The long-term goal of the research is to fully understand the human genetic variation for traits important for bacterial pathogenesis and how they impact human disease.
Lab Members
Publications
Representative Publications:
Wang L, Pittman KJ, Barker JR, Salinas RE, Stanaway IB, Williams GD, Carroll RJ, Balmat T, Ingham A, Gopalakrishnan AM, Gibbs KD, Antonia AL, Network Te, Heitman J, Lee SC, Jarvik GP, Denny JC, Horner SM, Delong MR, Valdivia RH, Crosslin DR, and Ko DC. (2018) An atlas of genetic variation for linking pathogen-induced cellular traits to human disease. Cell Host & Microbe. 24(2): 308-323. Web portal: http://h2p2.oit.duke.edu/
Bourgeois JS, Zhou D, Thurston TLM, Gilchrist JJ, and Ko DC. (2018) Methylthioadenosine suppresses Salmonella virulence. Infect Immun. 86 (9) e00429-18.
Jaslow SL, Gibbs KD, Fricke WF, Wang L, Pittman KJ, Mammel MK, Thaden JT, Fowler VG, Jr., Hammer GE, Elfenbein JR, and Ko DC. (2018) Salmonella Activation of STAT3 Signaling by SarA Effector Promotes Intracellular Replication and Production of IL-10. Cell Reports. 23(12):3525-36.
Alvarez MI and Ko DC. (2018) Reply to Gilchrist et al.: Possible roles for VAC14 in multiple infectious diseases. Proc Natl Acad Sci U S A. 115(16):E3604-E3605.
Alvarez MI, Glover LC, Luo P, Wang L, Theusch E, Oehlers SH, Walton EM, Tram TTB, Kuang YL, Rotter JI, McClean CM, Chinh NT, Medina MW, Tobin DM, Dunstan SJ, and Ko DC. (2017) Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1706070114. PubMed PMID: 28827342.
Wang L, Ko ER, Gilchrist J, Pittman KJ, Rautanen A, Pirinen M, Thompson JW, Dubois LG, Langley RG, Jaslow SL, Salinas RE, Rouse DC, Moseley A, Mwarumba S, Njuguna P, Mturi N, Wellcome Trust Case-Control Consortium 2, The Kenyan Bacteraemia Study Group, Williams TN, Scott AG, Hill AVS, Woods CW, Ginsburg GS, Tsalik EL and Ko DC. (2017) Human genetic and metabolite variation reveal methylthioadenosine is a prognostic biomarker and inflammatory regulator in sepsis. Science Advances. 3(3):e1602096.
Galarza-Muñoz G, Briggs FBS, Evsyukova I, Bergamaschi L, Wang L, Ko DC, Bradrick SS, Barcellos LF, Garcia-Blanco MA, and Gregory SG. (2017) The RNA helicase DDX39B regulates IL7R splicing reducing the risk of Multiple Sclerosis. Cell. 169(1):72-84.e13.
Pittman KJ, Glover LC, Wang L, and Ko DC. (2016) The Legacy of Past Pandemics: Common Human Mutations that Protect Against Infectious Disease. PLoS Pathogens. 12(7):e1005680.
Oehlers SH, Flores MV, Hall CJ, Wang L, Ko DC, Crosier KE, Crosier PS.(2016) A whole animal chemical screen approach to identify modifiers of intestinal neutrophilic inflammation. FEBS J. doi:10.1111/febs.13976 [Epub ahead of print].
Wang L, Oehlers SH, Espenschied ST, Rawls JF., Tobin DM, and Ko DC. (2015) CPAG: software for leveraging pleiotropy in GWAS to reveal similarity between human traits links plasma fatty acids and intestinal inflammation. Genome Biology. 16:190.
Salinas RE, Ogohara C, Shukla KP, Thomas MI, Miller SI, and Ko DC. (2014) A cellular genome-wide association study reveals human variation in microtubule stability and a role in inflammatory cell death. Mol Biol Cell. 25(1):76-86.
Ko DC, Urban TJ. (2013) Understanding human variation in infectious disease susceptibility through clinical and cellular GWAS. PLoS Pathogens. 9(8):e1003424.
Ko DC, Gamazon ER, Shukla KP, Pfuetzner RA, Whittington D, Holden TD, Brittnacher MJ, Fong C, Radey M, Ogohara C, Stark AL, Akey JM, Dolan ME, Wurfel MM, Miller SI. (2012) Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. PNAS. 109:E2343-2352.
Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, Vary JC, Dunstan S, Farrar JJ, Thwaites G, King MC, Serhan CN, Ramakrishnan L. (2012) Therapies directed to LTA4H genotype can optimize the inflammatory response to mycobacterial infections. Cell. 148(3):434-46.
Fong C, Ko DC, Wasnick M, Radey M, Miller SI, Brittnacher M. (2010) GWAS Analyzer: integrating genotype, phenotype and public annotation data for genome-wide association study analysis. Bioinformatics. 26(4):560-4.
Ko DC, Shukla KP, Fong C, Wasnick M, Brittnacher MJ, Wurfel MM, Holden TD, O’Keefe GE, Van Yserloo B, Akey JM, Miller SI. (2009) A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease. Am J Hum Genet. 85(2):214-27.
Ko DC, Milenkovic L, Beier S, Manuel H, Buchanan J, Scott MP. (2005) Cell-autonomous death of cerebellar Purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genetics. 1(1):e7.
Ko DC, Binkley J, Sidow A, Scott MP. (2003) The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci U S A. 100(5):2518-25.
Ko DC, Gordon MD, Jin JY, Scott MP. (2001) Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol Biol Cell. 12(3):601-14.
Bourgeois JS, Wang L, Rabino AF, Everitt J, Alvarez MI, Awadia S, Wittchen ES, Garcia-Mata R, and Ko DC. (2021) ARHGEF26 enhances Salmonella invasion and inflammation in cells and mice. PLoS Pathog. 17(7):e1009713. PMC8294491.
Wang L, Balmat TJ, Antonia AL, Constantine FJ, Henao R, Burke TW, Ingham A, McClain MT, Tsalik EL, Ko ER, Ginsburg GS, DeLong MR, Shen X, Woods CW, Hauser ER, and Ko DC. (2021) An atlas connecting shared genetic architecture of human diseases and molecular phenotypes provides insight into COVID-19 susceptibility. Genome Med. 13(1):83. PMC8127495. Web portal: http://cpag.oit.duke.edu.
Bourgeois JS, Smith CM, Ko DC. (2021) These Are the Genes You’re Looking For: Finding Host Resistance Genes. Trends Microbiol. 29(4):346-362. PMC7969353.
Schott BH, Antonia AL, Wang L, Pittman KJ, Sixt BS, Barnes AB, Valdivia RH, Ko DC. (2020) Modeling of variables in cellular infection reveals CXCL10 levels are regulated by human genetic variation and the Chlamydia-encoded CPAF protease. Sci Rep. 10(1):18269. PMC7588472.
Gibbs KD, Washington EJ, Jaslow SL, Bourgeois JS, Foster MW, Guo R, Brennan RG, and Ko DC. (2020) The Salmonella Secreted Effector SarA/SteE Mimics Cytokine Receptor Signaling to Activate STAT3. Cell Host Microbe. S1931-3128(19)30594-3. PMC6952535.
Antonia AL, Gibbs KD, Trahair ED, Pittman KJ, Martin AT, Schott BH, Smith JS, Rajagopal S, Thompson JW, Reinhardt RL, and Ko DC. (2019) Pathogen Evasion of Chemokine Response Through Suppression of CXCL10. Front Cell Infect Microbiol 9:280. PMC6693555.
For a complete list of publications, click here.
Data & Analysis Tools
Make a Gift
At Duke University School of Medicine, donors play a vital role in fostering innovation, enhancing patient care, and training the next generation of health care leaders. Your contributions not only sustain and elevate existing programs but also pave the way for groundbreaking discoveries.

You can support the work of the Department of Molecular Genetics and Microbiology by making a gift to the Dennis Ko Laboratory.






